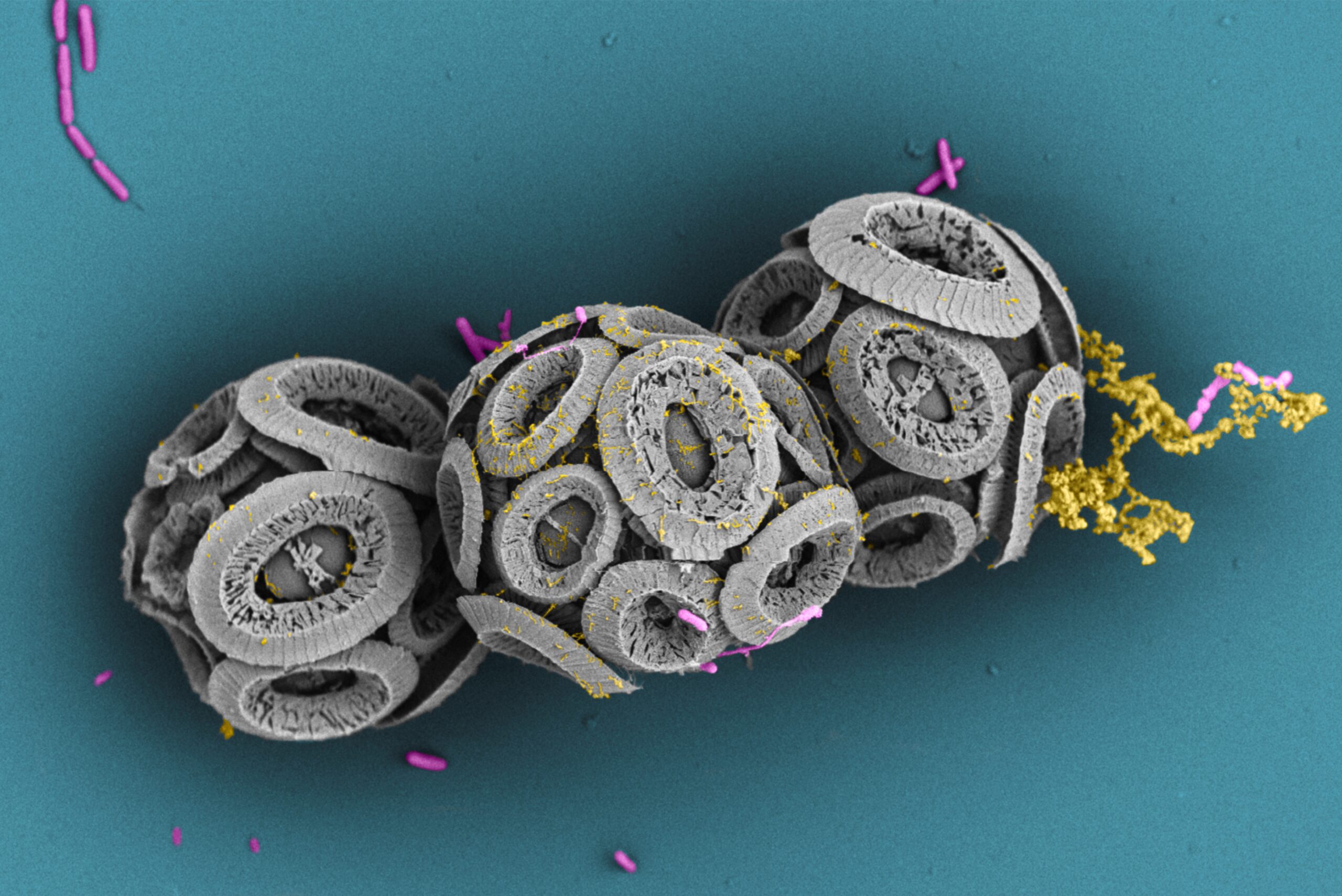
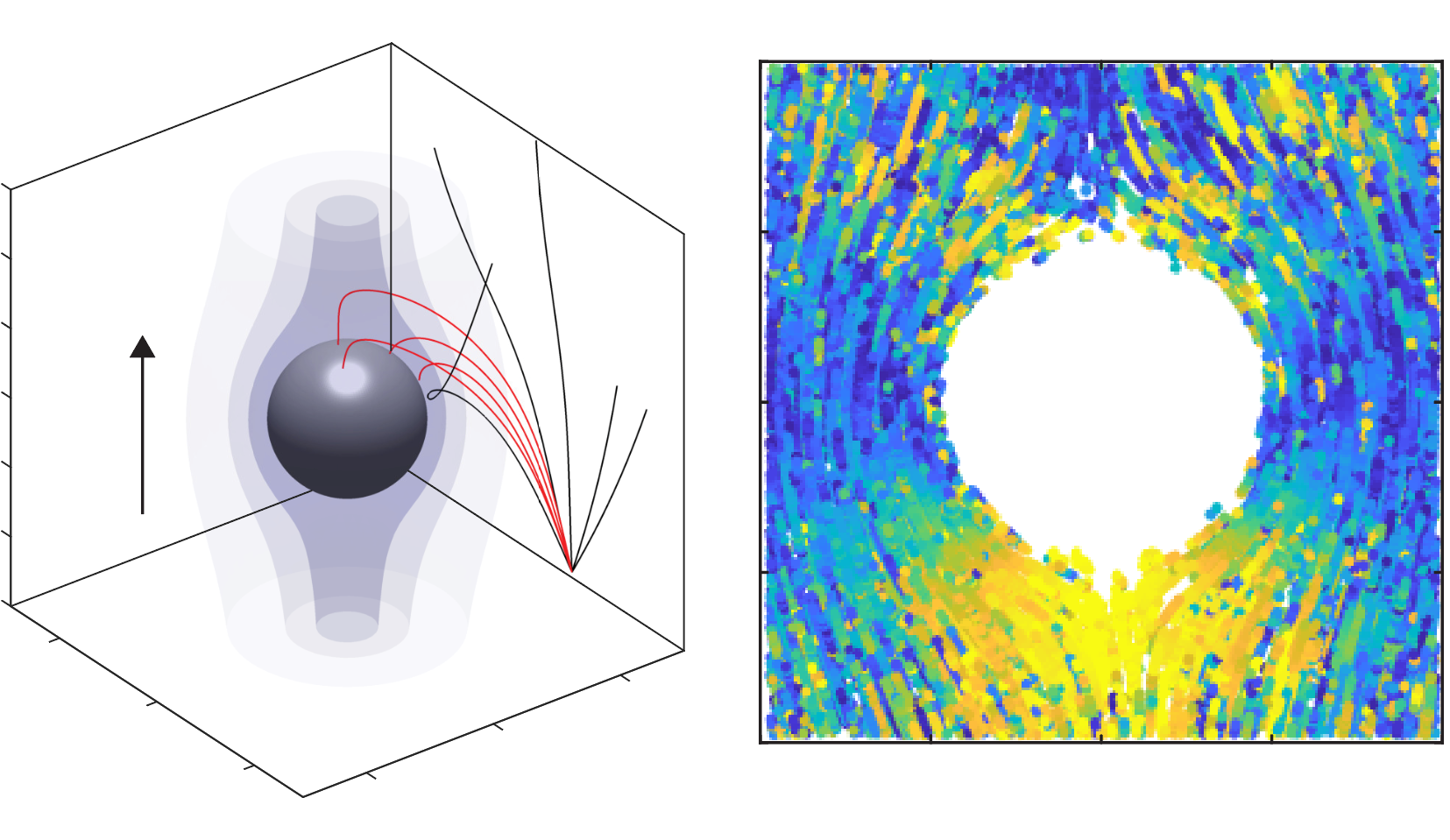
We study a range of environmental processes at the interface between engineering and ecology, to discover how they work and to determine their environmental and societal effects. Through our research, we aim to contribute to generating the knowledge that will help better understand and manage the environment, particularly in the face of ongoing climate change.
Bacterial motility & Chemotaxis
#chemotaxis, motility, ocean, biological carbon pump, symbiosis, microfluidics, microscopy, field deployments
Microbial Ecosystems
#microbial ecology, environmental microbiology, population dynamics, microbial growth scaling law