Biophysics of bacteria-phytoplankton interactions
Contact: Riccardo Foffi, Dr. Jonasz Słomka
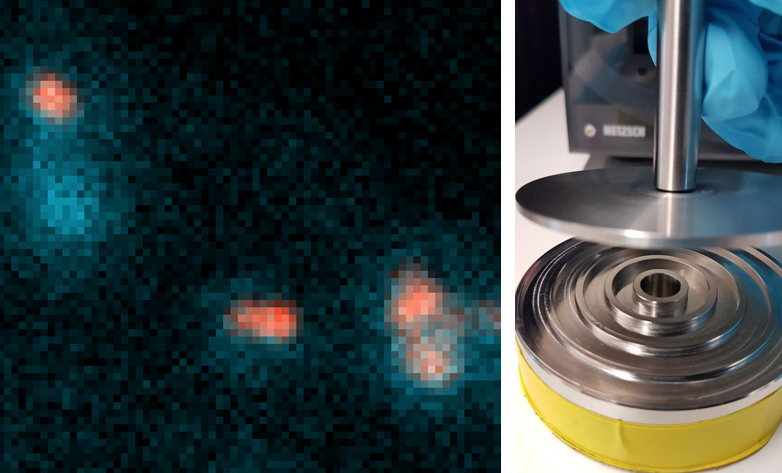
Perform agent-based simulations of bacteria-phytoplankton systems, investigating how different physical properties (bacterial speed, swimming strategy, phytoplankton shape and size…) modulate uptake and interaction rates. There is flexibility for the student to suggest complementary studies.
How often do bacteria exchange genes in aquatic environments?
Contact: Dr. Jonasz Słomka
Completion of the development of a method to measure the frequency of bacterial conjugation events as a function of the conjugation rate and its application in varying conditions.
Software development for automated study of freshwater ecology
Contact: Dr. Luiz Martins da Silva
Calibrate the novel iteration of the Riverine Organism Drift Imager (RODI) and develop a script in Python to select only images of interest for recording to extend the autonomous field deployment time.
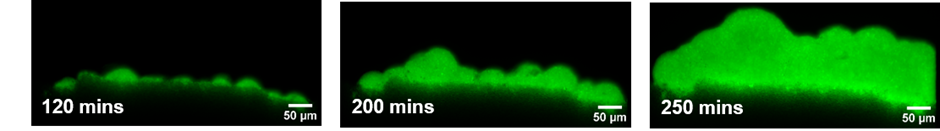
A microfluidic investigation of spatial arrangement of microorganisms in a biofilm for sustainable light-driven wastewater treatment
Contact: Dr. Eleonora Secchi
The aim of this project is to study the relationship between mass transfer in the porous biofilm matrix and the location of the different species within the biofilm. The fundamental understanding gained in this work will later be used to optimize the growth of phototrophic species and biofilm formation in a photobioreactor, allowing sustainable wastewater treatment.
Control and systems development of a real-time microfluidic antibiotic screening and single-cell detection platform
Contact: Dr. Sam Charlton
The student will have access to the components of the system, our image analysis library and mechanical test protocols currently validated in the lab. The student will not be expected to perform any wet-lab work. This project is aimed at students with an interest in designing systems for biotechnology and bio-manufacturing applications.
Exploring the ecological role of bacterial attachment in facilitating bacteria–phytoplankton interaction
Contact: Dr. Mathieu Forget
The goal of this project is to gain a better understanding of the diversity of mechanisms and the ecological roles of bacterial attachment in bacteria–phytoplankton associations. Students will be encouraged to propose new approaches to investigate specific research directions related to the project, based on their interests. Depending on the student’s background and motivation, this experimental approach can be complemented by numerical simulations.
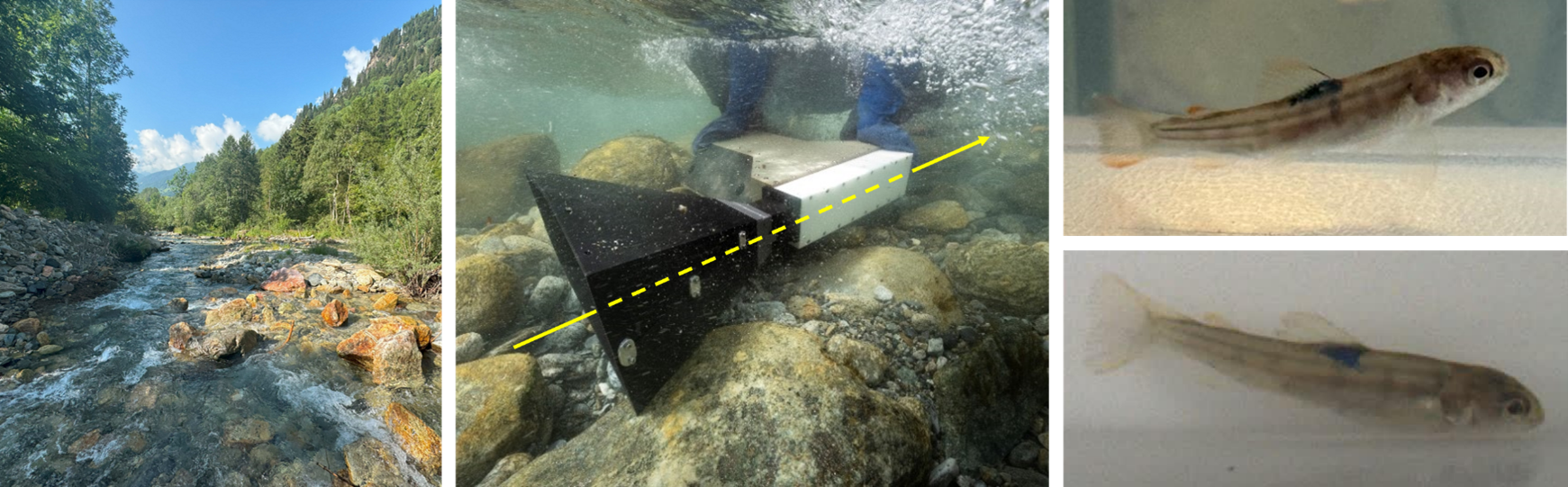
Quantifying the effects of hydropeaking on the dispersal of brown trout larvae (Salmo trutta) in the Poschiavino river (GR)
Contact: Joël Wittmann, Dr. Luiz Silva
The aim of the Master Thesis or Project is to measure the dispersal of brown trout larvae and identify differences between reaches affected by hydropeaking and residual flow reaches.
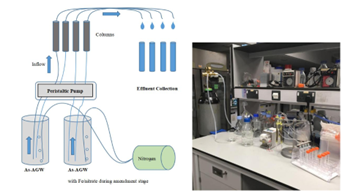
Numerical modeling of column experiments to study arsenic immobilization in groundwater from Yinchuan Basin, China
Contact: Prof. Joaquin Jimenez-Martinez (jjimenez@ethz.ch)
Perform numerical simulations of arsenic immobilization to represent column experiments and use numerical modeling to investigate different in-situ remediation approaches to mitigate arsenic contamination in groundwater.